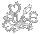Drosera arcturi, Photo © Richard Nunn.
The evolution of Drosera and the other genera in the Droseraceae provides as many twists and turns in the story as any Gothic novel. It all begins in the dim past, an era so long ago we may never be able to know exactly what happened. We have self-incompatibility which in itself merits its own page and changes in chromosome numbers also with a separate discussion. Hybridization between species may be lurking in the closets. On this page we look at what the DNA is telling us. Chloroplast DNA provides simple answers but does not tell the whole story. Nuclear DNA may say something completely different while hybridization, polyploidy, and random chance muddy the waters. In the end we have a huge diversity of Drosera species in a large extended family.
Why is the evolution of Drosera important? Yes it is fun to know the evolutionary history of your plants. But what is important is to understand evolution of plants in general. We are in the early throws of a major mass extinction on this planet. Yes, it is important to know what we are losing, but it is even more important to understand how the species came into being. We know many of the workings of evolution in plants and we are constantly finding out more about plant biology and evolution. But finding new evolutionary mechanisms and determining which evolutionary mechanisms the plants actually experienced will tell us much more about how life came to be as it is. Or was.

Drosera arcturi. Drosera arcturi and its close relative Drosera murfetii appear to have maintained the expected ancestral leaf form of the genus. Notice how the leaf base wraps the stem of the plant in a way similar to Dionaea, Nepenthes, and other related genera. Photo © Denis Barthel / Wikimedia Commons / CC-BY-SA-3.0 / GFDL.
This figure was constructed using data from NCBI and aligned by clustalw-2.0.10, analyzed by PHYLIP 3.69 dnaml DNA maximum likelihood program, graphed via PHYLIP drawtree unrooted tree drawing program and detailed manually in a drawing program. The lengths of the lines are an indication of genetic distance. Plumbago auriculata was used as the outgroup.
Most of the data used in this analysis are those used by Fernando Rivadavia, Katsuhiko Kondo, Masahiro Kato, and Mitsuyasu Hasebe (2003) which includes data in the NCBI database from other authors. The species identifications are supplied by the persons submitting the data to NCBI. There are no vouchers or other source documentation required by NCBI so the data may be misidentified. Not all data used here are published in journals.
Drosera spatulata (1) sequence is from Williams, S. E., Chase, M. W. and Albert, V. A. NCBI accession L19530 (2000). There is no information about the source or publication.
Drosera spatulata (2) sequence is from Hoshi, Y., Shirakawa, J. and Hasebe, M. NCBI accession AB355695 (2007). The source location is Japan.
2x and 4x refer to the ploidy level of lineages. 2x is diploid. 4x is tetraploid.
The chloroplast rbcL sequence data clearly delineate the Drosera out-of-Australia hypothesis. Drosera may not have initially evolved in Australia, but the genus certainly diversified dramatically there. One early branch of the Drosera tree gave rise to the specialized tuberous, woolly, and pygmy sundews along with their relatives Drosera binata and Drosera glanduligera. The other branch gave rise to or continued the more generalized small rosetted sundews with Drosera burmannii, Drosera adelae, Drosera hamiltonii, Drosera indica, and Drosera stenopetala. A few of the these Australian species established limited populations in Asia, India, and Africa. There are also examples of two species being established in South America from close relatives in Australia / New Zealand. However it appears the vast majority of Drosera species on the continents other than Australia originated relatively recently (i.e. a few tens of millions of years) from a plant similar to what we know today as Drosera spatulata.
As discussed by Rivadavia, et. al (2003) it appears the diploid Drosera in North America and South America arrived from a migration out of Australia up eastern Asia, across to North America, and then to South America. There are some curious anomalies though. The Australian and Asian Drosera spatulata extant today as well as Drosera neocaledonica are tetraploids. The only known diploid Drosera spatulata are in New Zealand. Also with the data we have now, Drosera brevifolia from the southern USA is more basal than Drosera rotundifolia from just north of Japan—the source location for the plant sequenced. We do not know specifically about Drosera rotundifolia from North America and Europe and it may be different from its Asian populations. In Asia we could be seeing the result of extinction or replacement or hybridization of the more ancestral forms. Once this branch of Drosera reached South America there is evidence of exchange back and forth between North America and South America. This is not surprising because there is a major bird migration route where these species are found. Birds were probably also responsible for the migration up Asia and over to North America as well.
What is difficult to explain is the migration route for the tetraploid Drosera from Australia or Asia to South America and Africa. Those continents appear to have been colonized at approximately the same time and many of the Drosera there now look very similar to each other and to Drosera spatulata. A problem of determining a source location is the known basal tetraploids are in Australia and Asia. One possibility is there was a tetraploid Drosera spatulata-like plant in Antarctica before it froze over with the opening of the land bridge between Antarctica and South America 25 million years ago. Birds transported the seeds north into South America and Africa. Alternatively the tetraploid Drosera spatulata in Australia and/or Tasmania could have traveled by sea birds on the prevailing winds to the southern tip of South America and Africa or via ocean currents. Drosera sessilifolia is too closely related to Drosera burmannii to have taken the Antarctica route so transportation half way around the world does happen.
Besides the Drosera spatulata nexus there is another nexus point that needs study: the initial Droseraceae radiation. The placement of Dionaea / Aldrovanda / Drosera regia / Drosera arcturi on the nexus of the rbcL tree and the chloroplast matK tree (See Nepenthes Evolution) are the same and not exactly satisfying. Based on characters other than DNA, Drosera regia and Drosera arcturi are transitional between Dionaea and the rest of the Drosera species, so some confusion is expected. The chloroplast-based trees indicate Drosera regia is off the main Drosera branch and more closely related to Aldrovanda. Or the flip side of this is Aldrovanda is more closely related to Drosera regia than it is to Dionaea. How much of what we see in the trees is real and how much is an artifact of the data or how we do the analysis? Never mind we are asking questions about events that happened more than 50 million years ago. Drosera regia and Dionaea muscipula appear to be allohexaploids, Aldrovanda vesiculosa an allo-octoploid and Drosera arcturi an allotetraploid (see Drosera Chromosomes page for an explanation). Since the rbcL tree only tracks chloroplast lineages, it only shows one of the possible three or four species that contributed to allopolyploids we see today. Even looking at nuclear DNA may not give a satisfying answer because diploidization could have silenced or removed the other copies of the sequences. In other words we may never know how all these species are related. But we can still try to find out.
The program used to generate the rbcL tree above uses an algorithm to determine which tree is the one most likely tree. There are programs that can tell you how confident they are at various points in the tree. The usual response is to collapse low confidence branch points and just say we do not know. What you really want to know is why there is low confidence, what are the consequences of that low confidence, and it would be nice to see visually what the range of alternative trees look like.
The NeighborNet algorithm (Bryant and Moulton 2004) takes a different approach to displaying species relationships. It draws a relationship network rather than forcing the data into a strict single line, bifurcating tree structure. It does this by first doing a maximum likelihood tree then layering a maximum parsimony tree on top. If the data display as tight lines we have good confidence in the groupings. If the data take the form of a vast network of connections then we have problems. In a chloroplast gene it is assumed problems causing low confidence are related to data quality. Low quality can result from sequencing issues, too few diagnostic sites, and/or a high ratio of within species changes to between species changes. The Drosera chloroplast data suffer from the latter two issues. Also since the rbcL sequences are for a region that produces a protein there is the additional problem of back-mutations. That is, in a lineage there may be a change to an amino acid in the protein that may not be optimal. Later in the lineage it may mutate back to the original protein. In the programs doing the analysis this mutation and back-mutation would be consider no mutation when in fact it is two mutation events. Unfortunately there is no way effectively deal with this in a proper scientific way because there is no way to know this has happened until after the analysis is finished. And then you do not really know for sure. At least with NeighborNet you can see evidence of this happening because the distances along the tree are not additive and NeighborNet takes that into account.

Drosera murfetii is a very close relative of Drosera arcturi. Photo © Richard Nunn.

Drosera brevifolia from the SE USA.

White flowered variant of Drosera spatulata var. gympiensis found along the coast near Gympie, Queensland, Australia. Photo by Ivan Snyder.

Typical Drosera spatulata from Queensland, Australia. This is also known as the 'Kanto' form.
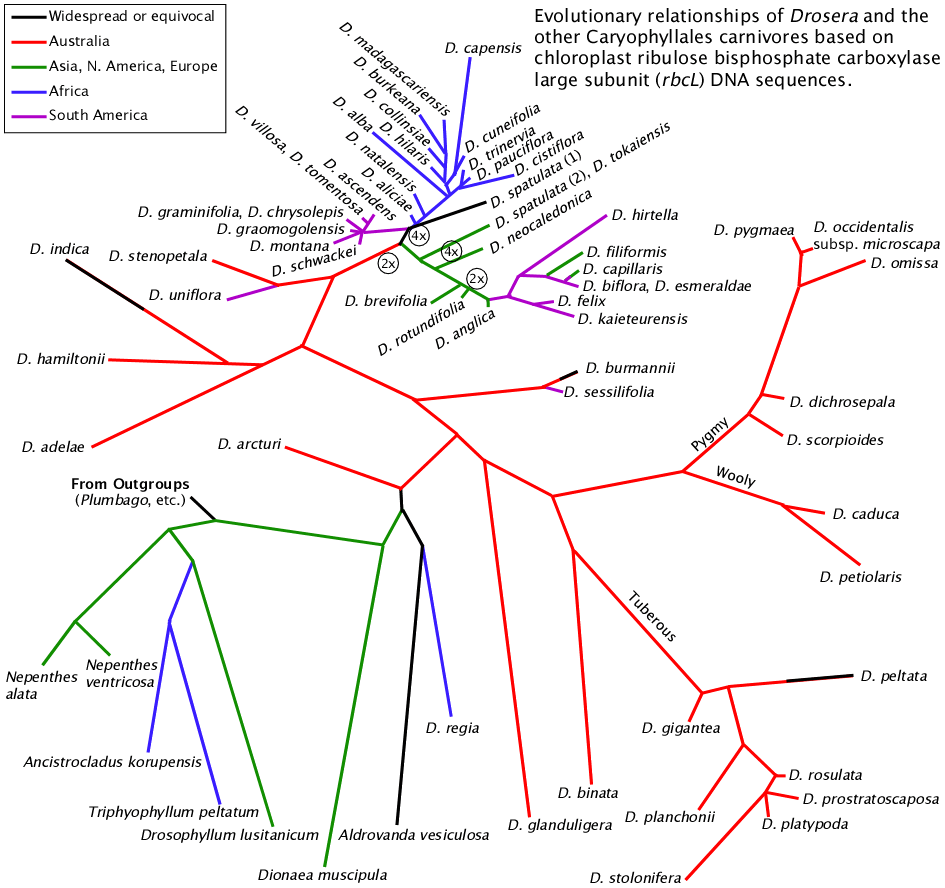
NeighborNet relationship network of the same chloroplast rbcL data set used for the tree above. The data were analyzed and drawn by the SplitsTree4 program (Drosera H. Huson and Drosera Bryant, Application of Phylogenetic Networks in Evolutionary Studies, Mol. Biol. Evol., 23(2):254-267, 2006) using the default settings. Changing program parameters made no significant changes in the network.
The NeighborNet tree clearly illustrates the problems at the two major nexus points. It shows the data do not allow a clear delineation of Drosera regia, Drosera arcturi, Aldrovanda vesiculosa, and Dionaea muscipula at the Droseraceae radiation. And again it favors Aldrovanda closer to Drosera regia and Drosera arcturi than to Dionaea. This is not totally unexpected because there is evidence each of those species evolved their signature attributes independently. Aldrovanda appears to have evolved snap traps independently of Dionaea: the traps of Aldrovanda close using a hinge-like mechanism while the traps of Dionaea close by modifying the curvature of the leaf. While Drosera regia has many attributes that ally it with the core Drosera it does have a few traits more like those of Dionaea than the rest of the Drosera genus. In fact we have no idea what any of these species looked like 65 million years ago or when each key character evolved.
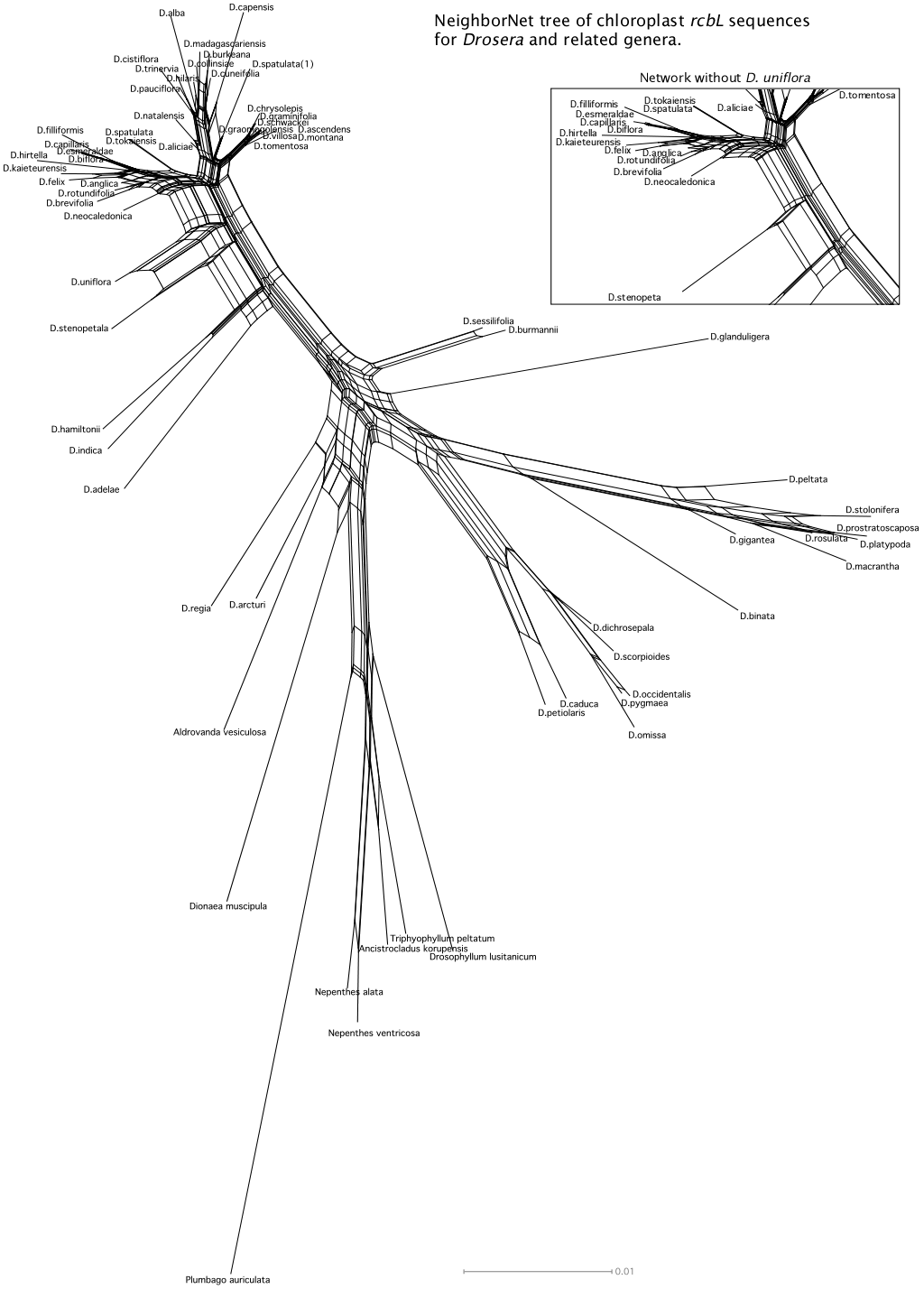
More DNA data may help us to understand the Droseraceae radiation but maybe not. The figure to the right is a network of the combined rbcL and matK sequences for the species where we have adequate data. Basically it says the same thing. The most likely explanation for the difficulty is when Drosera regia, Drosera arcturi and the other Drosera, Aldrovanda vesiculosa, and Dionaea muscipula diverged the crucial period of time may have been a small fraction of the time period since then. If all five proto-species in the Droseraceae radiation diverged within, say, a million years and this happened 70 million years ago there will be very few diagnostic loci and we may not be able to distinguish diagnostic loci from noise.
The out-of-Australia nexus also shows a lot of what could be considered unresolved connections. This nexus may suffer from some of the same issues as the initial Droseraceae radiation—the key events happened over a short period of time a long time ago. However in this case it is not as long ago and we have more species with sequence data and many more species, subspecies, and forms that could be sequenced. Working with the data we have, removing Drosera uniflora from the analysis removes much of the network between Drosera stenopetala / Drosera uniflora and Drosera neocaledonica. Something curious is going on there but what? The non-linearity in the relationships makes me wonder if Drosera has low level chloroplast transfer in pollen. However it is more likely that with more data Drosera uniflora may link in more closely with another species closer to the branch point. But which species? It may no longer be around. On the South America / Africa branch we see that what I have labeled "Drosera spatulata (1)" sits right in this branch point and it networks curiously to Drosera capensis. I would sure like to know more about the source plant for that sequence.
It is important to note the data used to make the trees are chloroplast lineages. Chloroplasts are essentially intracellular cyanobacteria symbionts. Their genetics is much simpler which makes the data cleaner and thus easier to analyze and interpret. This chloroplast sequences also changes more slowly than most nuclear sequences and at a rate that is good for this level of study. What can complicate the analysis is that the rbcL sequence is of a protein coding region. Only certain mutations can happen that still result in a functional enzyme. Within a lineage there can be mutations and then back-mutations to the previous state obscuring the actual history of the lineage.
If there is a hybridization event, the chloroplasts will generally come from the female parent. For instance we know that Drosera tokaiensis resulted from a cross between the diploid (2x) Drosera rotundifolia and a tetraploid (4x) Drosera spatulata. This gave a mostly sterile triploid (3x) which after rare chromosome doubling resulted in a fertile hexaploid (6x) Drosera tokaiensis with the Drosera spatulata chloroplasts. Something similar probably happened to make the tetraploid (4x) Drosera spatulata found in Asia. I suspect a diploid (2x) Drosera spatulata hybridized with a diploid (2x) species currently identified as "Drosera spatulata" but not or a now extinct species which after chromosome doubling resulted in the tetraploid (4x) "Drosera spatulata" with Drosera spatulata chloroplasts. Another example of this is the hybridization between Drosera rotundifolia and Drosera linearis to produce Drosera anglica. At this point it is not clear whether Drosera anglica got its chloroplasts from Drosera rotundifolia and the event happened long ago or whether it has Drosera linearis chloroplasts. There is no published Drosera linearis chloroplast sequence.
What is a problem for the phylogeny is multiple chloroplast lineages in a species. If populations of a species have been isolated long enough the chloroplast profiles in each population will evolve independently. This is because new chloroplast lineages are founded by mutations all the time. There is generally no mixing or recombination of lineages in individuals because of the exclusive maternal inheritance. Through random sampling or selection, different lineages can come to dominate in separate populations of the same species. Hybridization between interfertile species can also inject new chloroplast lineages into the mix. This can make things "interesting" and some of our conclusions wrong. Unfortunately at this point we do not know how common these exceptions are.
How can we determine both parents of a hybridization event? With the chloroplast sequence data we can determine the probable female parent of the hybrid if it happened relatively recently. If the hybridization event happened multiple times we should be able to find some plants with chloroplasts from one parent and other plants with chloroplasts of the other parent. That is of course if the hybridization works both ways which is not always the case.
Another way to look at hybridization events is to compare nuclear DNA sequences. A common sequence to use is the DNA sequence for ribosomal RNAs and the "spacer" sequences between them. In the case for Drosera tokaiensis, Hoshi et al. (2008) found the nuclear sequences to be the same as Drosera rotundifolia. Curiously they did not find any sequences that matched Drosera spatulata. However they did find sequences that do not match any published sequence. It is unclear at this point whether they found evidence of gene silencing or if there were DNA isolation and sequencing issues. Either way they did confirm that Drosera tokaiensis is of hybrid origin and the parents were Drosera spatulata as the seed parent and Drosera rotundifolia as the pollen parent for the samples tested.
This Drosera story is a work in progress. We may never know or understand all the twists and turns but as we get more data more of the story will become less opaque.
-- John Brittnacher
January 2011
For a more detailed discussion please see the following articles and articles they reference.
Rivadavia, Fernando, Katsuhiko Kondo, Masahiro Kato, and Mitsuyasu Hasebe (2003) Phylogeny of the sundews, Drosera (Droseraceae) based on chloroplast rbcL and nuclear 18S ribosomal DNA sequences. American Journal of Botany 90(1): 123-130.
Bryant, Drosera , and V. Moulton. (2004) NeighborNet: an agglomerative algorithm for the construction of planar phylogenetic networks. Mol. Biol. Evol. 21:255–265.
Nakano, M., E. Kinoshita and K. Ueda. (2004) Life history traits and coexistence of an amphidiploid, Drosera tokaiensis, and its parental species, Drosera rotundifolia and Drosera spatulata (Droseraceae). Plant Species Biology 19(2):59–72.
Snyder, Ivan (2003) Curious Natural Hybrid Sundews. Carniv. Pl. Newslett. 32(2):52-56. ( PDF )
Snyder, Ivan (2000) Colchicine Treatment on Sterile Hybrid sundews. Carniv. Pl. Newslett. 29(1):4-10. ( PDF )
Poppinga, Simon and Marc Joyeux (2011) Different mechanics of snap-trapping in the two closely related carnivorous plants Dionaea muscipula and Aldrovanda vesiculosa. Physical Review E 84, 041928.
The NCBI sequences used in this analysis are from:
Rivadavia, Fernando, Katsuhiko Kondo, Masahiro Kato, and Mitsuyasu Hasebe (2003) Phylogeny of the sundews, Drosera (Droseraceae) based on chloroplast rbcL and nuclear 18S ribosomal DNA sequences. American Journal of Botany 90(1): 123-130.
Hoshi, Y., Shirakawa, J., Hasebe, M., Fukushima, K., and Kondo, K. (2008). Tandem repeat rDNA sequence derived from parents were stably maintained in hexaploids of Drosera spathulata complex (Droseraceae). Cytologia 73: 313-325.
Ichihashi,Y. and Minami,M.
Chubu University, Japan
Unpublished data
Albert,V. A., Williams,S.E. and Chase,M. W.
Lebanon Valley College, PA, USA
Science 257 (5076), 1491-1495 (1992)
Cameron,K. M., Wurdack,K. J. and Jobson,R. W.
The New York Botanical Garden, NY, USA
Am. J. Bot. 89 (9), 1503-1509 (2002)
NeighborNet network for combined chloroplast rbcL and matK data sets. The data are from NCBI, aligned by clustalw-2.0.10, and analyzed and drawn by the SplitsTree4 program. The combined data sets do not help resolve the initial Droseraceae radiation.
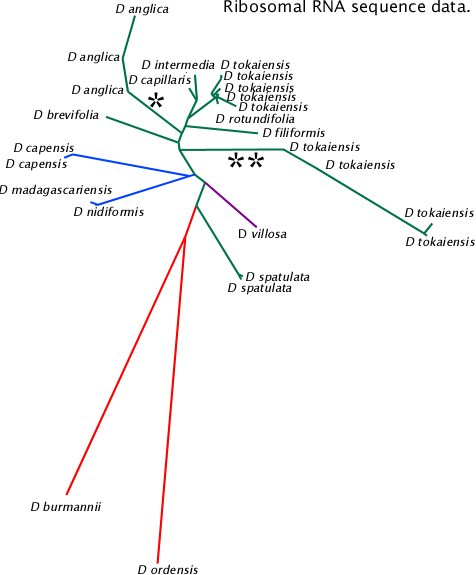
Associations between 18S rRNA, ITS1, 5.8S rRNA, ITS2 and 28S rRNA sequences in NCBI database. The tree data were generated by NCBI Blast and graphed with PHYLIP drawtree. This tree is based on shorter sequences than the NeighborNet network below.
The lines marked ![]() and
and ![]()
![]() are species of apparent hybrid origin. It is not known if the different sequences result from legitimate or illegitimate recombination events or laboratory errors.
are species of apparent hybrid origin. It is not known if the different sequences result from legitimate or illegitimate recombination events or laboratory errors.
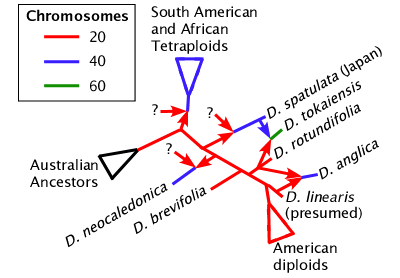
None of the bifurcating phylogeny trees, including NeighborNet, can actually represent the true nature of the Drosera phylogeny. We do not know how many species are the result of hybridization that did not result in a change in chromosome number. However we do know which species are allopolyploids. In allopolyploids the genomes of two species are combined but the chloroplast lines are not. The South American and African Drosera derived from a plant similar to Drosera spatulata are all allopolyploids, usually allotetraploids. Drosera spatulata in Japan may be from the same source but is more likely a separate polyploidization event. Drosera neocaledonica, Drosera tokaiensis, and Drosera anglica are all independently derived allopolyploids. As in the case with Drosera tokaiensis you can see that allopolyploids are not necessarily derived from very closely related species.